UK Biobank, a large-scale biomedical database and research resource, has made public data from 45,000 Magnetic Resonance Imaging (MRI) scans analysed by a team of University of Westminster researchers, including Professor Louise Thomas, Professor Jimmy Bell, Dr Nicolas Basty and Dr Brandon Whitcher.
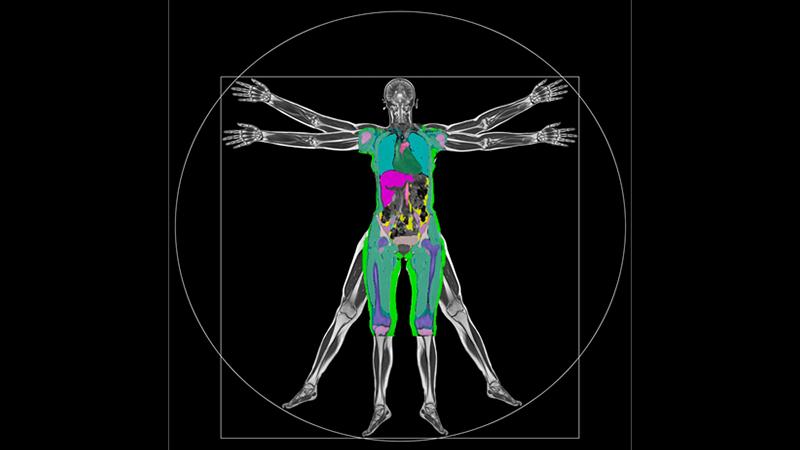
The data is the result of an ongoing collaboration between the University and scientists from Calico Life Sciences LLC, through which they developed a deep-learning (DL) based pipeline that automatically analyses MRI scans, including the data of over 40,000 individuals so far. DL is an AI powered machine learning method that can create multiple layers of algorithms and networks, making large scale data processing more efficient.
The UK Biobank research have captured a vast number of MRI scans of the human body together with genetic and lifestyle data. The study aims to improve understanding of why certain people develop life-altering diseases when others do not, enabling scientists to diagnose, treat, and potentially prevent diseases like dementia, heart disease, arthritis and cancer.
The data generated by Westminster and Calico includes a number of clinically relevant image-derived-phenotypes (IDPs): liver and pancreas volume, and their fat and iron content, along with volumes of lung, spleen, kidney, and subcutaneous and visceral fat volume. The researchers have also returned the first of the repeated imaging datasets, so long-term changes in participants can now be tracked.
The research started back in 2021, when a study of 38,000 abdominal MRI scans by Westminster and Calico, in cooperation with Biobank UK, was published in the renowned scientific journal eLife. This study demonstrates the advantage of DL, in quantifying and systemising health parameters from MRI across a range of organs and tissues, and to generate new insights into the genetic structure of these traits.
An analysis pipeline and all related programs and codes are now publicly available to anyone interested in setting up their own analysis pipeline.
Talking about the research, Louise Thomas, Professor of Metabolic Imaging and Study Investigator at the University of Westminster, said: “The work we have undertaken at Westminster in partnership with our collaborators at Calico LLC over the last few years developing the pipeline to automatically develop this data has been an extraordinary advance. Manually creating a dataset of this magnitude would be impossible. And this is a testament to the dedication and creativity of an outstanding research team. We are delighted that the UK Biobank have made our data publicly available and look forward to seeing how researchers across the world integrate it into their own research.”
The study was supported by Westminster’s Research Centre for Optimal Health (ReCOH), which was established to deepen the understanding of lifestyle-accelerated ageing, and to deliver solutions through this knowledge to support optimal health practices in adult life.
Learn more about ReCOH at the University of Westminster.


